Cancer Immune Escape and Transplantation Immunology
2. Transplantation immunology:
Transplantation has become a common surgical procedure that replaces patient organs that are damaged or ill functioning so they no longer sufficiently support the patient’s health and survival. Unfortunately, the immunosuppression needed to prevent the transplant rejection response by the immune system is associated with higher risks of sepsis and cancer. Thus, how can we minimize the effects of the immunosuppressive therapy currently needed? To approach this problem, we are taking our cues from exceptional patients who develop so-called “operational” tolerance towards their immunologically mismatched organ transplants, even though they have stopped taking immunosuppressive drugs. Our goal is therefore to find a set of genes that can induce this special immunological state of tolerance towards mismatched cells and organs and then translate our findings into localized immunomodulation therapy.
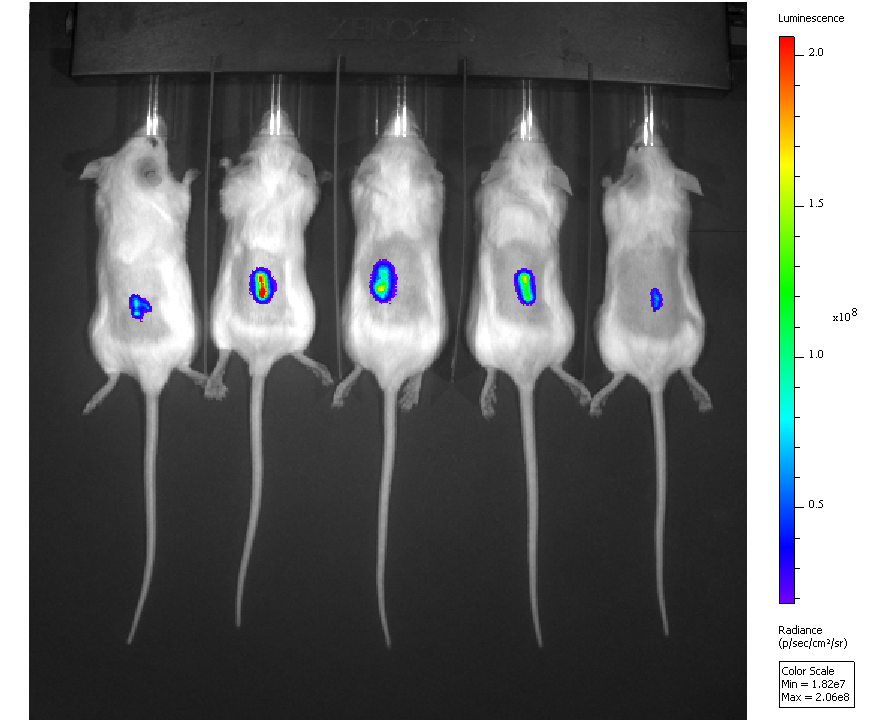
For both of these questions, we are investigating candidate genes in mouse models of disease for their causal role. Genes are either transferred in vitro into cells of interest for overexpression, or knocked out using mobile genetic elements called ‘transposons’. The gene-modified cells are subsequently introduced into mice where we then monitor the cells’ survival in real time using bioluminescence imaging (BLI). Cells with prolonged survival can then be recovered and analyzed for overexpressed or knocked out genes. Finally, in the case of cancer, we are also analyzing RNA sequence data of mouse tumors and publically available human cancer genomics databases to confirm the significance of the identified genes as contributors to disease progression.
1. Cancer Immune Escape
How do cancer cells escape from surveillance by the immune system that normally removes such altered cells? So far, a number of genes, signaling molecules and pathways have been implicated, yet, it remains unclear what minimal set of changes is sufficient to achieve this capability in cancer cells. Once we know this and fully understand the cancer immune escape mechanisms more effective immunotherapies – which are currently revolutionizing cancer treatment – can be designed so that these are no longer fraught with dangers, such as severe autoimmunity, and in some cases unfortunately, faster disease progression.
Many of the genes that drive the ten hallmarks of cancer (see Hanahan and Weinberg, Cell, 2000, 2011) have been identified except for those driving two of the hallmarks: the evasion from the immune system and chronic inflammation. The latter is largely determined by exogenous factors (ranging from physical agents and chemical toxins to infectious microbes).
To identify immune escape genes and pathways we are using a reverse genetics approach and mouse tumor cells that do not escape the immune system of wild type mice but do make tumors in immunodeficient mice (i.e. have all of the other hallmarks of cancer cells). We transfer individual and combinations of candidate genes into these cells and then inject them into immunocompetent mice where we observe them in real time using in vivo bioluminescence imaging (see image on the left). We then look for cells that live longer than control tumor cells and hence have escaped, with the aim of identifying the altered genes for this shift in phenotype.
In our preliminary studies we have used non-viral gene transfer to transfer 1) nine immunomodulatory genes commonly associated with immune escape, and 2) three strong cancer driver genes (oncogenes) - mutant K-ras also recently associated with immune escape, c-myc, and bcl-2 - in two-fold combinations into the immuno-sensitive tumor cells. When injected into immunocompetent mice, however, none of the cells survived longer than control cells. Thus, neither the combinations of the nine immunomodulatory genes nor the three oncogenes were sufficient to enable immune evasion. Therefore, wider genetic screen is needed.